Transfer learning methods for individualized treatment rules
Faculty Mentor: Shu Yang
Prerequisites: Probability, statistical computing.
Outline: The project is motivated by the 21st-century cures act, which emphasizes (i) harnessing real-world data for data-driven evidence and (ii) precision medicine that aims at learning individualized treatment regimes, tailoring to individual patient’s characteristics. Heterogeneous data sources are becoming increasingly popular for estimating treatment effects and learning individualized treatment regimes. Importantly, parallel randomized controlled trial data and large observational data present complementary features (see a review paper [1]). For example, randomized controlled trial data are free of confounding due to randomization of treatment by design, while they may be small and less representative of the real-world patient population [2]. On the other hand, observational data contains rich information in the patient portfolios while they suffer from data quality issues such as confounding, missingness, etc [1].
Research objectives: This project will review and develop data integration methods for estimating treatment effects and interpretable individualized treatment regimes (ITRs, such as linear rules) that leverage the unique strengths of data sources [3–5]. Figure 1 (left) illustrates the concept of the true and reduced optimal ITRs in the population. Figure 2 (right) illustrates the bias of the learned IRT based on a sample that is not representative of the target population. The research goal is to implement transfer learning to recover the optimal ITRs from the biased sample.
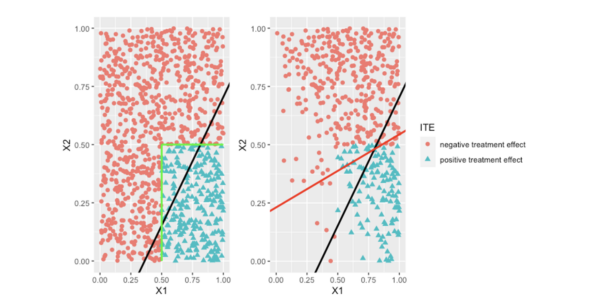
Figure 1. Illustration of true optimal ITRs. Every point represents the individual treatment effect value for individuals with (X1,X2): red dots mean negative treatment effects, and green triangles mean positive treatment effects. The left panel shows the true optimal ITR (X1>0.5 & X2<0.5, the green lines) that separates the positive treatment effects from the negative treatment effects and the population optimal linear ITR (the black line). The right panel shows the bias of the sample optimal linear ITR (the red line) when the sample is not representative of the target population.
Medical application: Transfer learning based on eICU-CRD and MIMIC-III data for treating patients with sepsis
We will conduct transfer learning using an application to data from the eICU collaborative research database (eICU-CRD) (Goldberger et al. 2000, Pollard et al. 2018, 2019) and the MIMIC-III clinical database (Goldberger et al. 2000, Johnson et al. 2016, 2019). Both MIMIC-III and eICU data consist of patients who suffered from sepsis. The eICU-CRD is a multi-center ICU database comprising de-identified health-related data associated with over 200,000 admissions to ICUs across the US between 2014-2015. The MIMIC-III database is a single-center ICU database comprising de-identified health-related data associated with over 40,000 patients who stayed in critical care units of the Beth Israel Deaconess Medical Center between 2001 and 2012. It is likely that the populations in the two databases have some heterogeneity. The goal is to conduct transfer learning of individualized treatment rules from one source population (e.g. eICU data source) to a target population (e.g. MIMIC-III data source).
Outcomes: The project will result in new methods for data fusion to improve treatment effect estimation.
References:
- Colnet B, Mayer I, Chen G, Dieng A, Li R, Varoquaux G, Vert J, Josse J, Yang S (2022). Causal inference methods for combining randomized trials and observational studies: a review, https://arxiv.org/abs/2011.08047v2.
- D. Lee, S. Yang, L. Dong, X. Wang, D. Zeng, J.W. Cai (2022). Improving trial generalizability using observational studies, Biometrics, doi:10.1111/biom.13609.
- L. Wu and S. Yang (2022). Integrative R-learner of heterogeneous treatment effects combining experimental and observational studies. CLeaR (Causal Learning and Reasoning) 2022.
- L. Wu and S. Yang (2021). Transfer learning of individualized treatment rules from experimental to real-world data, https://arxiv.org/abs/2108.08415.
- J. Chu, W. Lu, and S. Yang (2022). Targeted optimal treatment regime learning using summary statistics, https://arxiv.org/abs/2201.06229.

